Objectives: learn how to implement between subject mixture models (BSMM) and within subject mixture models (WSMM).
Projects: bsmm1_project, bsmm2_project, wsmm_project
Introduction
There exist several types of mixture models useful in the context of mixed effects models. It may be necessary in some situations to introduce diversity into the structural models themselves:
- Between-subject model mixtures (BSMM) assume that there exist subpopulations of individuals. Different structural models describe the response of each subpopulation, and each subject belongs to one of these subpopulations. One can imagine for example different structural models for responders, nonresponders and partial responders to a given treatment.
The easiest way to model a finite mixture model is to introduce a label sequence that takes its values in
such that
if subject i belongs to subpopulation m.
is the probability for subject i to belong to subpopulation m. A BSMM assumes that the structural model is a mixture of M different structural models:
$$f\left( t_{ij};\psi_i,z_i \right) = \sum_{m=1}^M 1_{z_i = m} f_m\left( t_{ij};\psi_i \right) $$
In other word, each subpopulation has its own structural model: is the structural model for subpopulation m.
- Within-subject model mixtures (WSMM) assume that there exist subpopulations (of cells, viruses, etc.) within each patient. In this case, different structural models can be used to describe the response of different subpopulations, but the proportion of each subpopulation depends on the patient.
Then, it makes sense to consider that the mixture of models happens within each individual. Such within-subject model mixtures require additional vectors of individual parameters representing the proportions of the M models within each individual i:
The proportions are now individual parameters in the model and the problem is transformed into a standard mixed effects model. These proportions are assumed to be positive and to sum to 1 for each patient.
Between subject mixture models
Supervised learning
- bsmm1_project (data = ‘pdmixt1_data.txt’, model = ‘bsmm1_model.txt’)
We consider a very simple example here with two subpopulations of individuals who receive a given treatment. The outcome of interest is the measured effect of the treatment (a viral load for instance). The two populations are non responders and responders. We assume here that the status of the patient is known. Then, the data file contains an additional column GROUP. This column is duplicated because Monolix uses it
- i) as a regression variable (X): it is used in the model to distinguish responders and non responders,
- ii) as a categorical covariate (CAT): it is used to stratify the diagnostic plots.
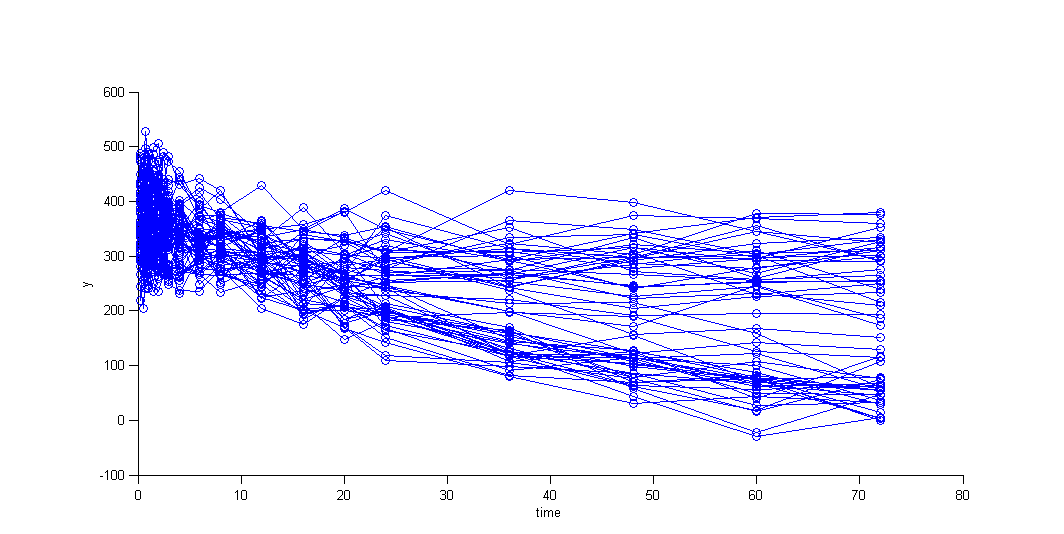
We can then display the data
and use the categorical covariate GROUP_CAT to split the plot into responders and non responders:
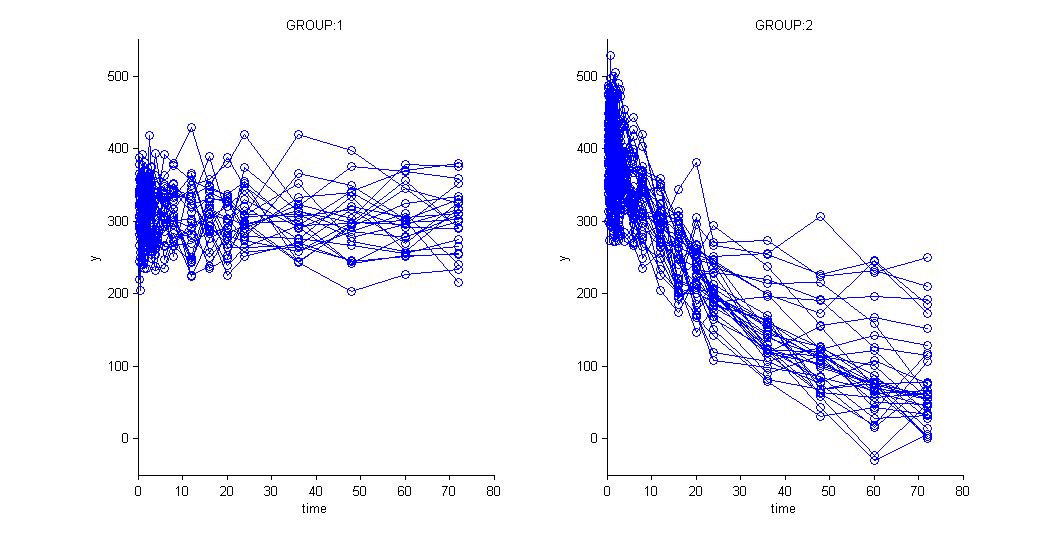
We use different structural models for non responders and responders. The predicted effect for non responders is constant:
while the predicted effect for responders decreases exponentially:
The model is implemented in the model file bsmm1_model.txt (remark that the names of the regression variable in the data file and in the model script do not need to match):
[LONGITUDINAL]
input = {A1, A2, k, g}
g = {use=regressor}
EQUATION:
if g==1
f = A1
else
f = A2*exp(-k*max(t,0))
end
OUTPUT:
output = f
The plot of individual fits exhibit the two different structural models:
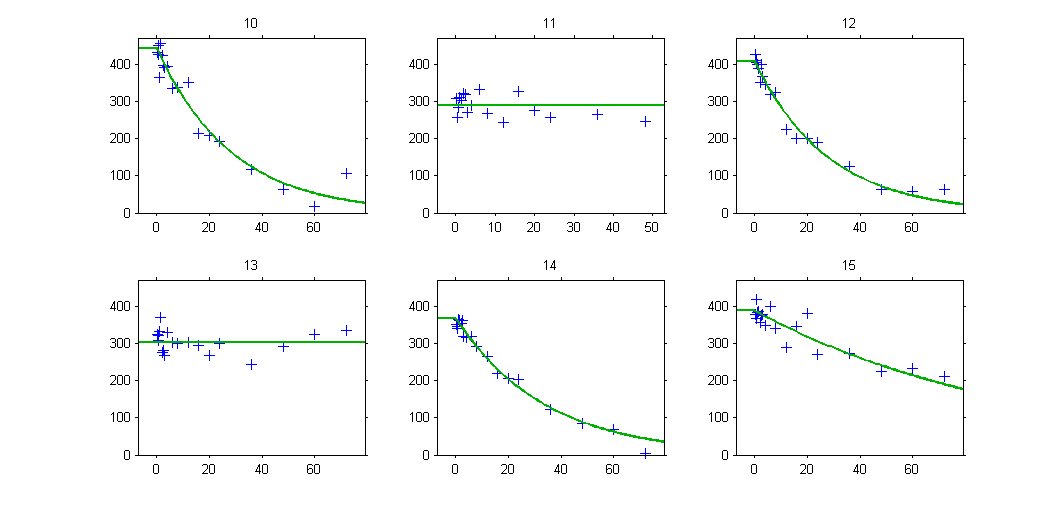
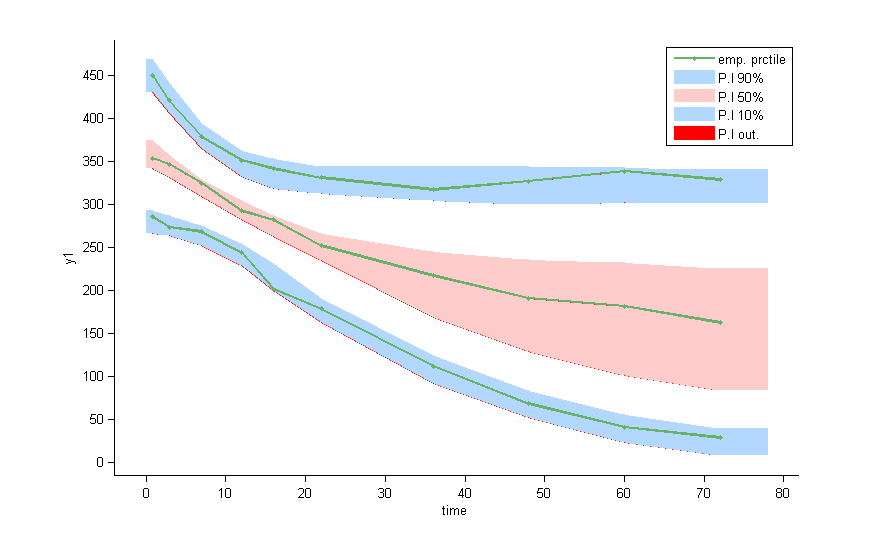
VPCs should then be splitted according to the GROUP_CAT
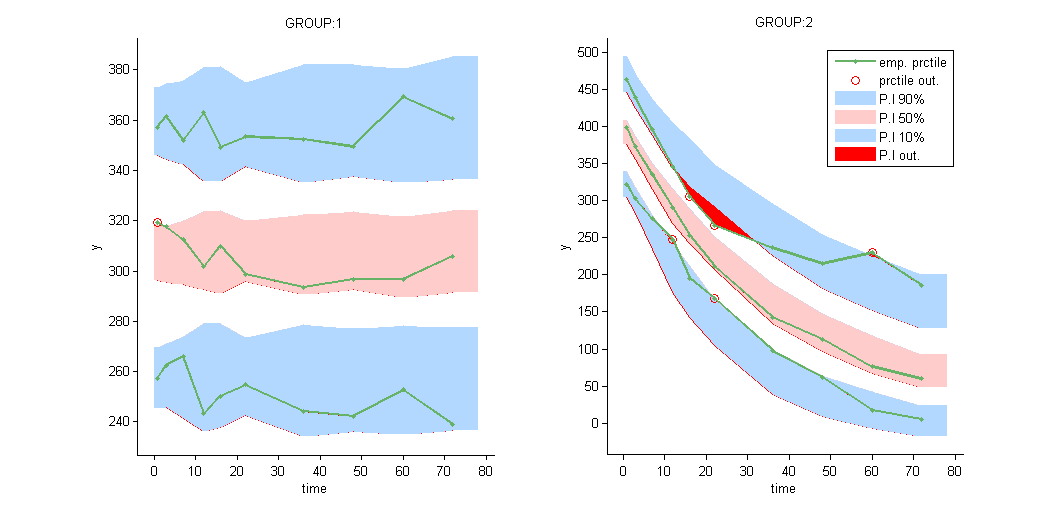
as well as the prediction distribution for non responders and responders:
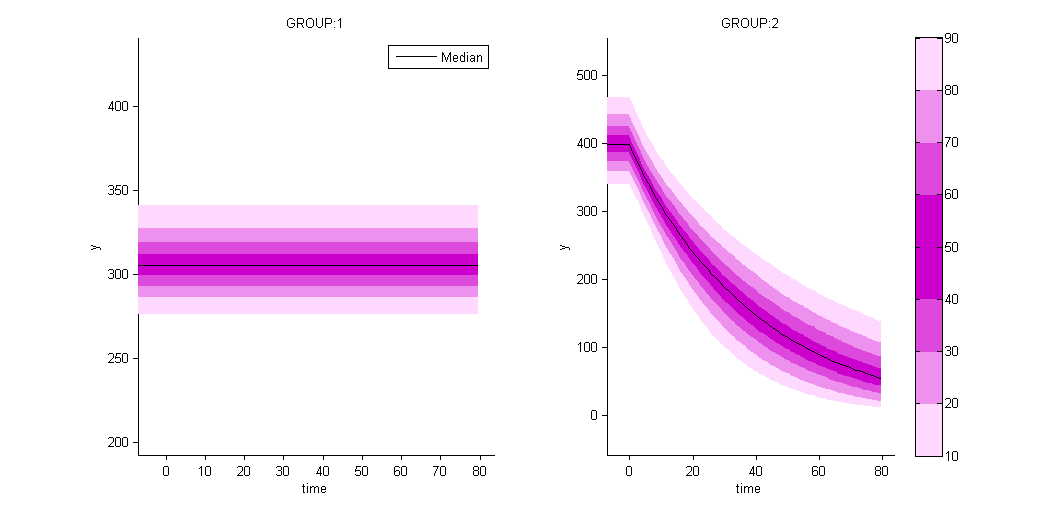
Unsupervised learning
- bsmm2_project (data = ‘pdmixt2_data.txt’, model = ‘bsmm2_model.txt’)
The status of the patient is unknown in this project (which means that the column GROUP is not available anymore):
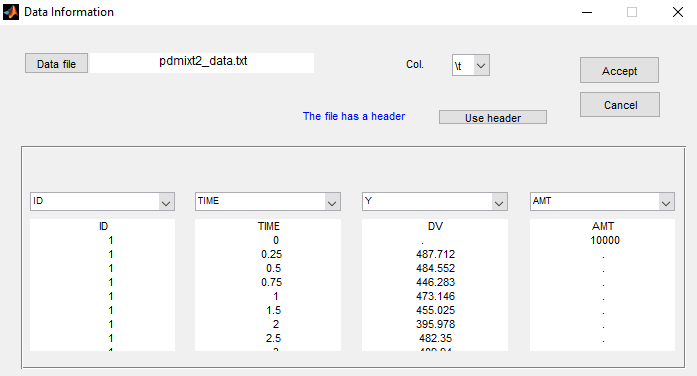
Let p be the proportion of non responders in the population. Then, the structural model for a given subject is with probability p and
with probability 1-p. The structural model is therefore a BSMM:
[LONGITUDINAL]
input = {A1, A2, k, p}
EQUATION:
f1 = A1
f2 = A2*exp(-k*max(t,0))
f=bsmm(f1, p, f2, 1-p)
OUTPUT:
output = f
Important: p is a population parameter of the model to estimate. There is no inter-patient variability on p: all the subjects have the same probability to be a non responder in this example. We use a logit-normal distribution for p in order to constrain it to be between 0 and 1, but without variability:
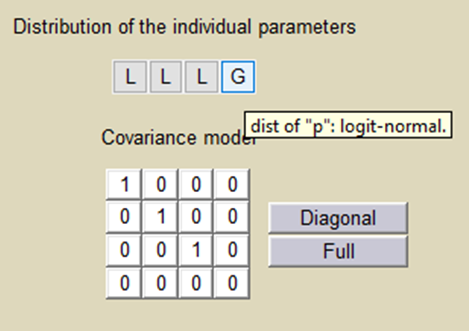
p is estimated with the other population parameters:
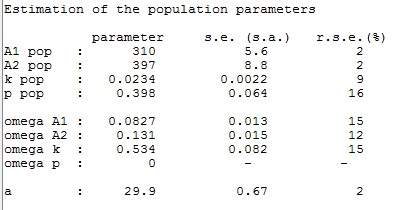
The individual parameters are estimated using the conditional modes in this project:
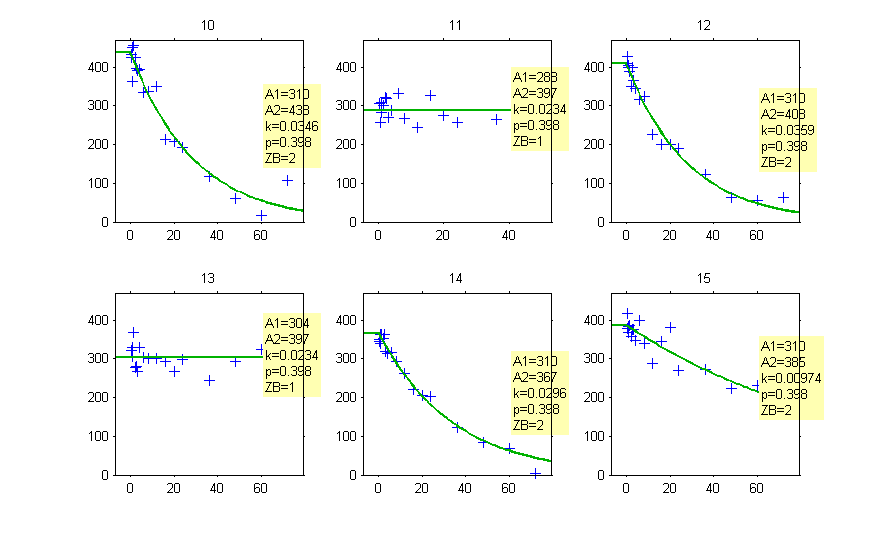
Then, the group to which a patient belong is also estimated as the group of highest conditional probability:
The estimated groups can be displayed together with the individual parameters:
and can be used as a stratifying variable to split some plots such as VPC’s
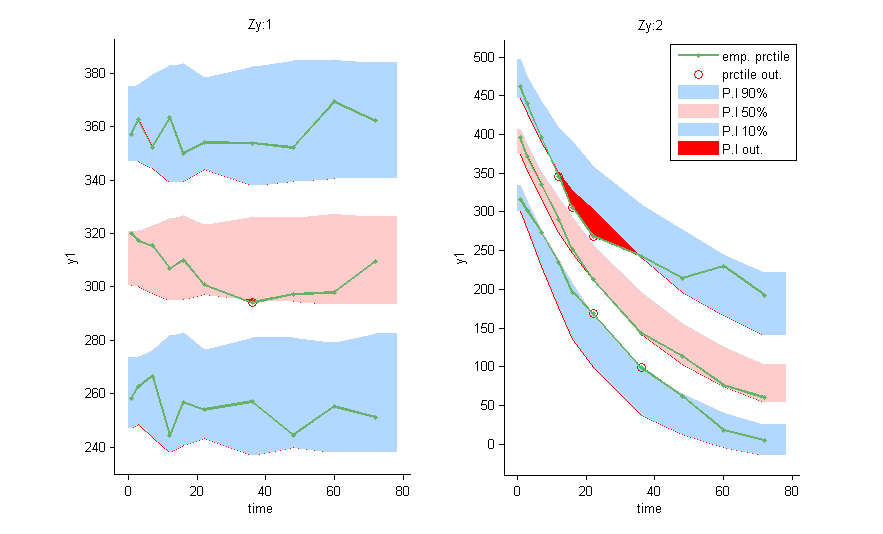
Within subject mixture models
- wsmm_project (data = ‘pdmixt2_data.txt’, model = ‘wsmm_model.txt’)
It may be too simplistic to assume that each individual is represented by only one well-defined model from the mixture. We consider here that the mixture of models happens within each individual and use a WSMM:
[LONGITUDINAL]
input = {A1, A2, k, p}
EQUATION:
f1 = A1
f2 = A2*exp(-k*max(t,0))
f=wsmm(f1, p, f2, 1-p)
OUTPUT:
output = f
Remark: Here, writing f=wsmm(f1, p, f2, 1-p) is equivalent to write f=p*f1 + (1-p)*f2
Important: Here, p is an individual parameter: the subjects have different proportions of non responder cells. We use a probit-normal distribution for p in order to constrain it to be between 0 and 1, with variability:
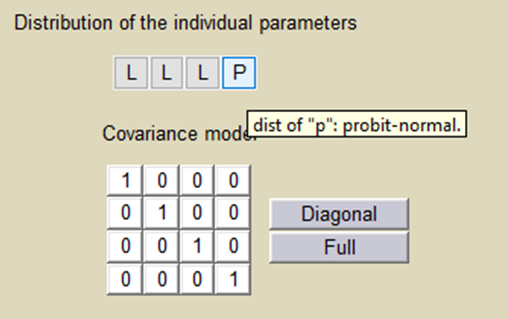
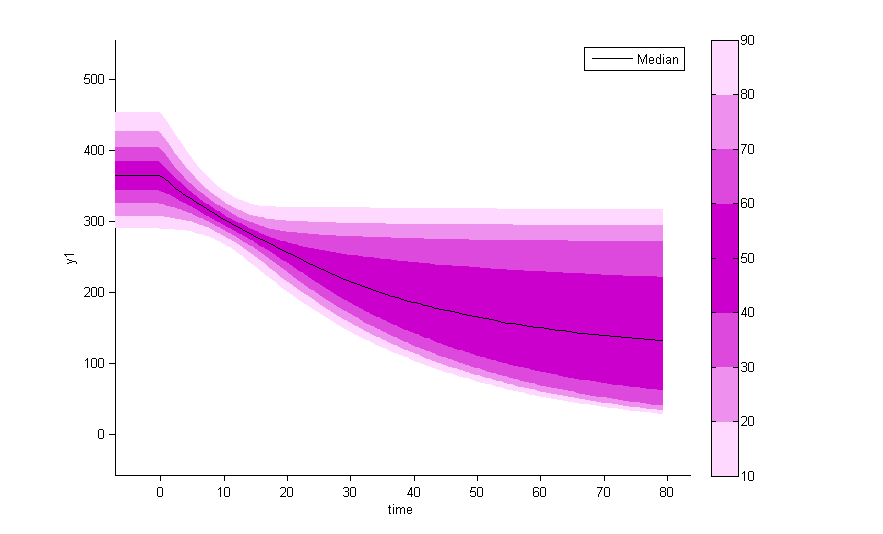
There is no latent covariate when using WSMM: mixtures are continuous mixtures. We therefore cannot split anymore the VPCs and the prediction distributions.