- Introduction
- Mixture of distributions – supervised learning
- Mixture of distributions – unsupervised learning
Objectives: learn how to implement a mixture of distributions for the individual parameters.
Projects: PKgroup_project, PKmixt_project
Introduction
Mixed effects models allow us to take into account between-subject variability. One complicating factor arises when data is obtained from a population with some underlying heterogeneity. If we assume that the population consists of several homogeneous subpopulations, a straightforward extension of mixed effects models is a finite mixture of mixed effects models, assuming, for instance, that the probability distribution of some individual parameters vary from one subpopulation to another one. The introduction of a categorical covariate (e.g., sex, genotype, treatment, status, etc.) into such a model already supposes that the whole population can be decomposed into subpopulations. The covariate then serves as a \emph{label} for assigning each individual to a subpopulation.
In practice, the covariate can either be known or not. Mixture models usually refer to models for which the categorical covariate is unknown, but whichever the case, the model is the same. The difference appears when having to perform certain tasks and in the methods needed to implement them. For instance, model building is different depending on whether the labels are known or unknown: we have supervised learning if the labels are known and unsupervised otherwise. For the sake of simplicity, we will consider a basic model that involves individual parameters and observations
. Then, the easiest way to model a finite mixture model is to introduce a label sequence
that takes its values in
such that
if subject i belongs to subpopulation m.
In some situations, the label sequence is known and can be used as a categorical covariate in the model. If
is unknown, it can modeled as a set of independent random variables taking its values in
where for
,
is the probability that individual i belongs to group m. We will assume furthermore that the
are identically distributed, i.e.,
does not depend on i for
.
Mixture of distributions – supervised learning
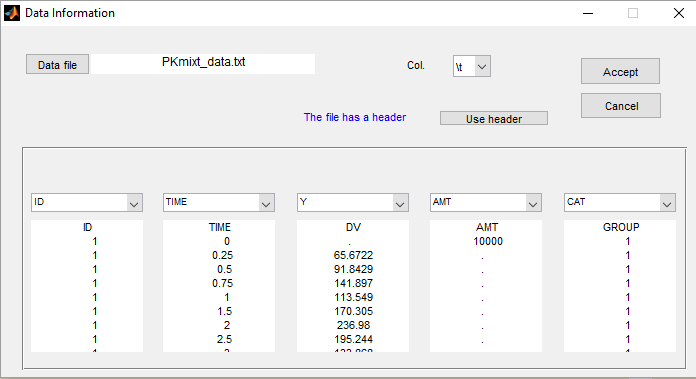
- PKgroup_project (data = ‘PKmixt_data.txt’, model = ‘lib:oral1_1cpt_kaVCl.txt’)
The sequence of labels is known as GROUP in this project and is therefore defined as a categorical covariate:
We can then assume, for instance different population values for the volume in the 2 groups:
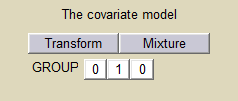
and estimate the population parameters using this covariate model
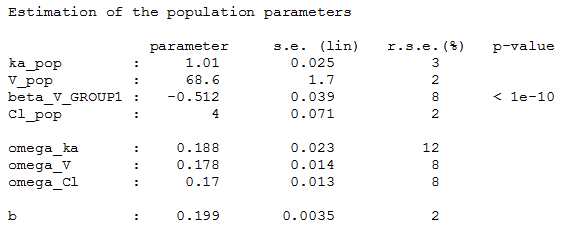
Then, this covariate GROUP can be used as a stratification variable.
Mixture of distributions – unsupervised learning
- PKmixt_project (data = ‘PKmixt_data.txt’, model = ‘lib:oral1_1cpt_kaVCl.txt’)
We will use the same data with this project but ignoring the column GROUP (which is equivalent to assume that the label is unknown)
If we suspect some heterogeneity in the population, we can introduce a “latent covariate” and select the number of categories for this latent covariate.
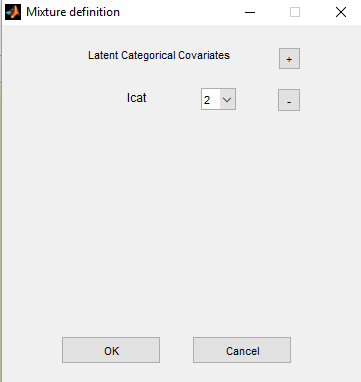
Remark: several latent covariates can be introduced in the model, with different number of categories.
We can then use this latent covariate lcat as any observed categorical covariate. We can assume again different population values for the volume in the 2 groups:
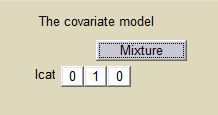
and estimate the population parameters using this covariate model. Proportions of each group are also estimated:
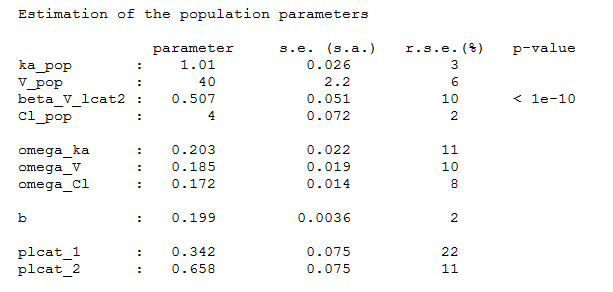
Once the population parameters are estimated, the sequence of latent covariates , i.e. the group to which belongs each subject, can be estimated together with the individual parameters, as the modes of the conditional distributions.
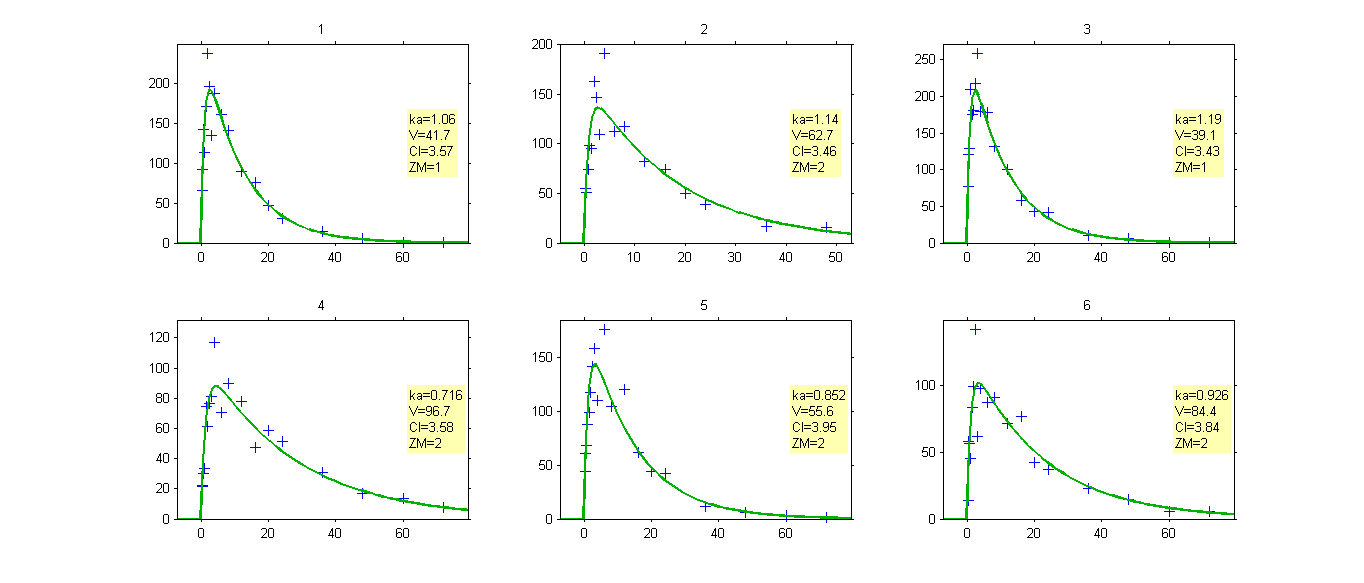
The sequence of estimated latent covariates can be used as a stratification variable:
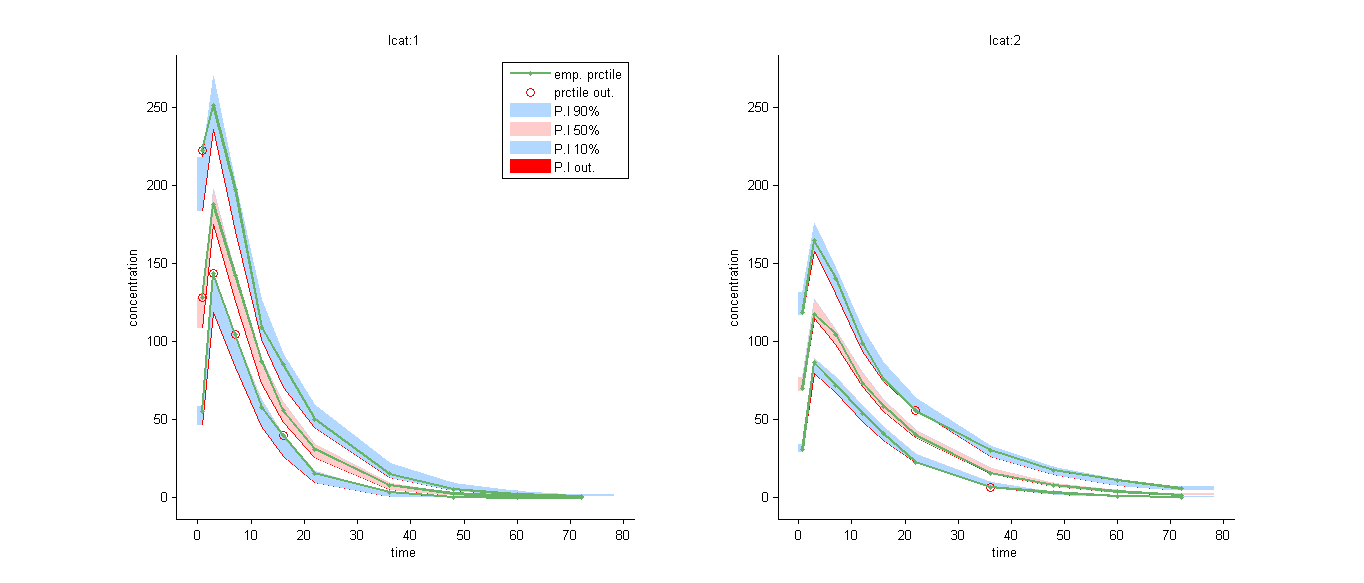
We can for example display the VPC in the 2 groups:
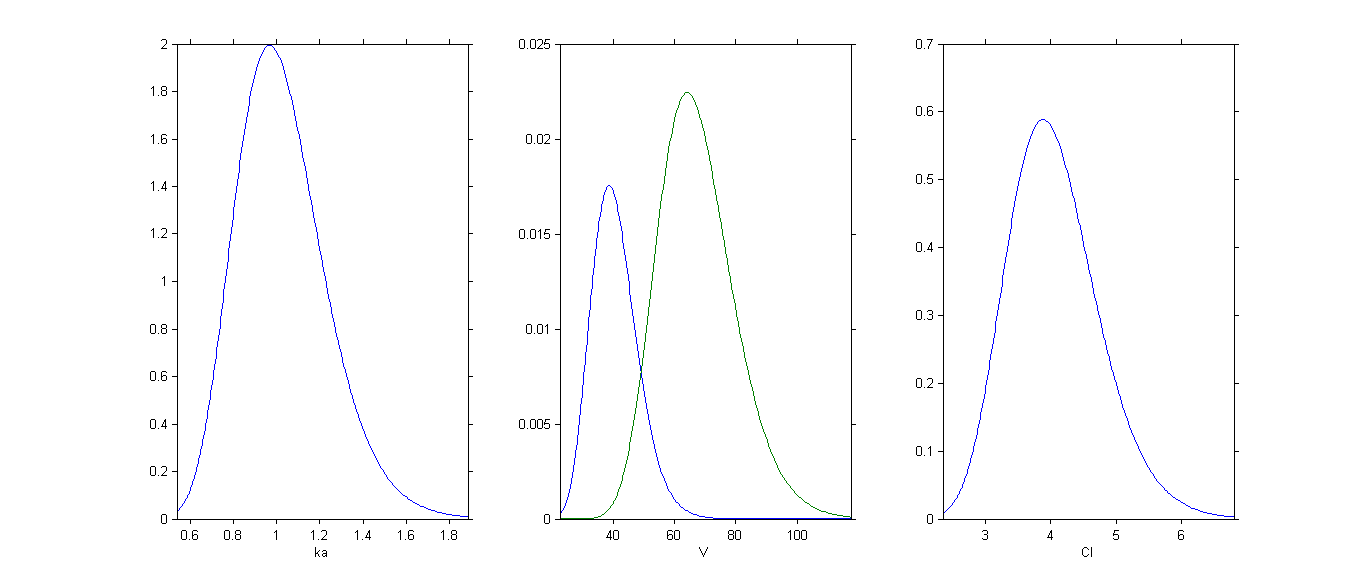
By plotting the distribution of the individual parameters, we see that has a bimodal distribution
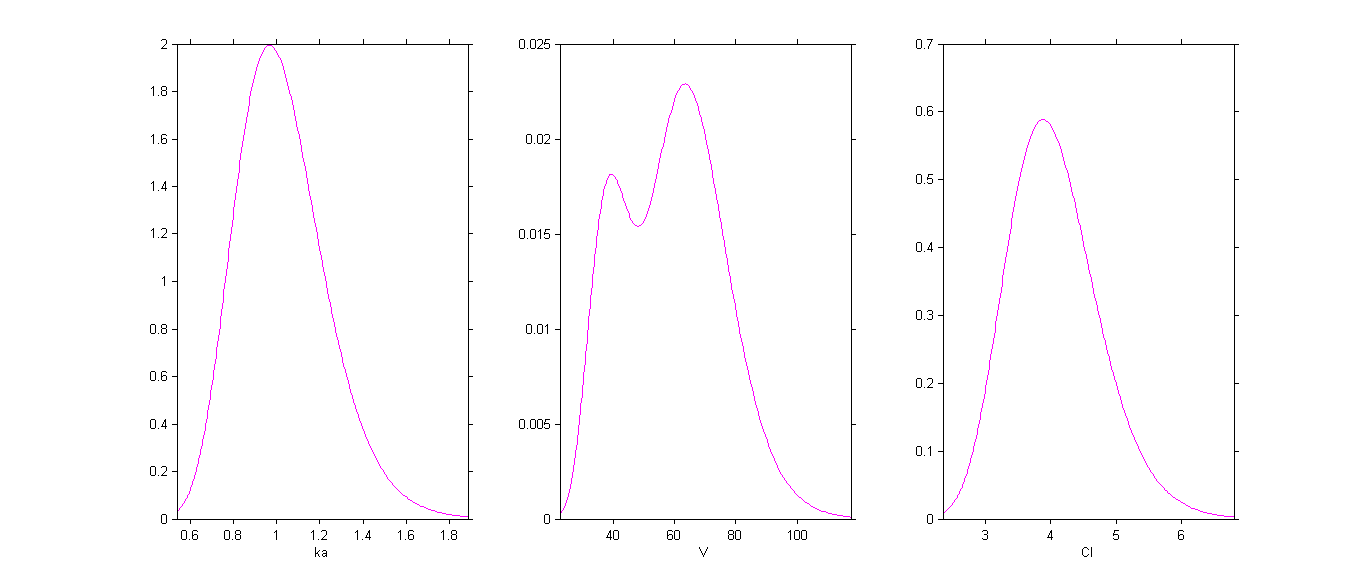
This distribution can be decomposed into 2 distributions by ticking the box By group: