- Introduction
- Marginal distributions of the individual parameters
- Correlation structure of the random effects
Objectives: learn how to define the probability distribution and the correlation structure of the individual parameters.
Projects: warfarin_distribution1_project, warfarin_distribution2_project, warfarin_distribution3_project, warfarin_distribution4_project
Introduction
One way to extend the use of Gaussian distributions is to consider that some transformation of the parameters in which we are interested is Gaussian, i.e., assume the existence of a monotonic function such that
is normally distributed. Then, there exists some
and
such that, for each individual
:
where is the predicted value of
. In this section, we consider models for the individual parameters without any covariate. Then, the predicted value of
is the
and
Transformation defines the distribution of
. Some predifined distributions/transformations are available in
Monolix:
- Normal distribution:
The two mathematical representations for normal distributions are equivalent:
- Log-normal distribution:
A log-normally random variable takes positive values only. A log-normal distribution looks like a normal distribution for a small variance . On the other hand the asymmetry of the distribution increases when
increases.
Remark: the two mathematical representations for log-normal distributions are equivalent:
- Power-normal (or Box-Cox) distribution:
Here, (with
) follows a normal distribution truncated so that
. It therefore takes positive values. This distribution converges to a log-normal one when
and a truncated normal one when
.
- Logit-normal distribution:
A random variable with a logit-normal distribution takes its values in
. The logit of
is normally distributed, i.e.,
- Probit-normal distribution:
A random variable with a probit-normal distribution also takes its values in (0,1).The probit function is the inverse cumulative distribution function (quantile function)
associated with the standard normal distribution
:
The probit-normal distribution with and
is the uniform distribution on
. Logit and probit transformations can be generalized to any interval (a,b) by setting
where
is a random variable that takes values in (0,1) with a logit-normal (or probit-normal) distribution.
Marginal distributions of the individual parameters
- warfarin_distribution1_project (data = ‘warfarin_data.txt’, model = ‘lib:oral1_1cpt_TlagkaVCl.txt’)
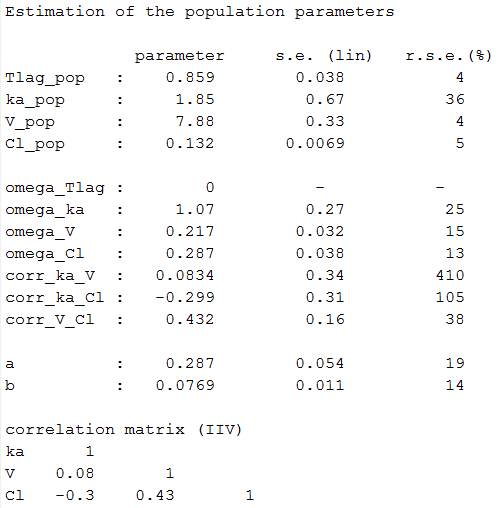
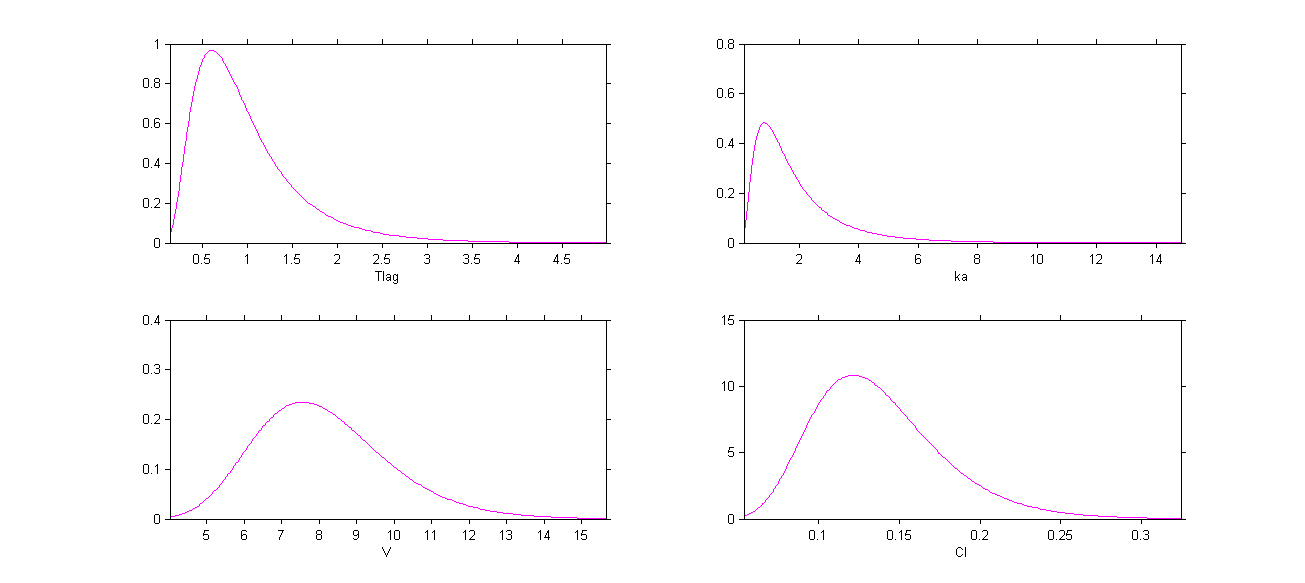
We use the warfarin PK example here. The four PK parameters Tlag, ka, V and Cl are log-normally distributed:
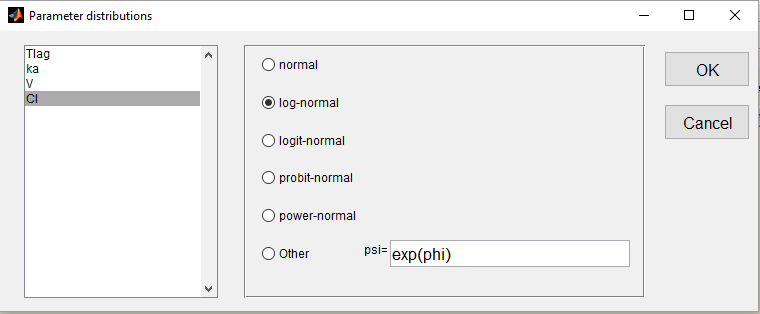
Letter L is then used for these four log-normal distributions in the main Monolix graphical user interface:
The distribution of the 4 PK parameters defined in the MonolixGUI is automatically translated into Mlxtran in the project file:
[INDIVIDUAL]
input = {Tlag_pop, omega_Tlag, ka_pop, omega_ka, V_pop, omega_V, Cl_pop, omega_Cl}
DEFINITION:
Tlag = {distribution=lognormal, typical=Tlag_pop, sd=omega_Tlag}
ka = {distribution=lognormal, typical=ka_pop, sd=omega_ka}
V = {distribution=lognormal, typical=V_pop, sd=omega_V}
Cl = {distribution=lognormal, typical=Cl_pop, sd=omega_Cl}
Estimated parameters are the parameters of the 4 log-normal distributions and the parameters of the residual error model:
Here, and
means that the estimated population distribution for the volume is:
or, equivalently,
where
.
Remark: is not the population mean of the distribution of
, but the median of this distribution.
The four probability distribution functions are displayed Figure Parameter distributions:
Click on Settings and select the checkbox Informations to display a summary of these distributions:
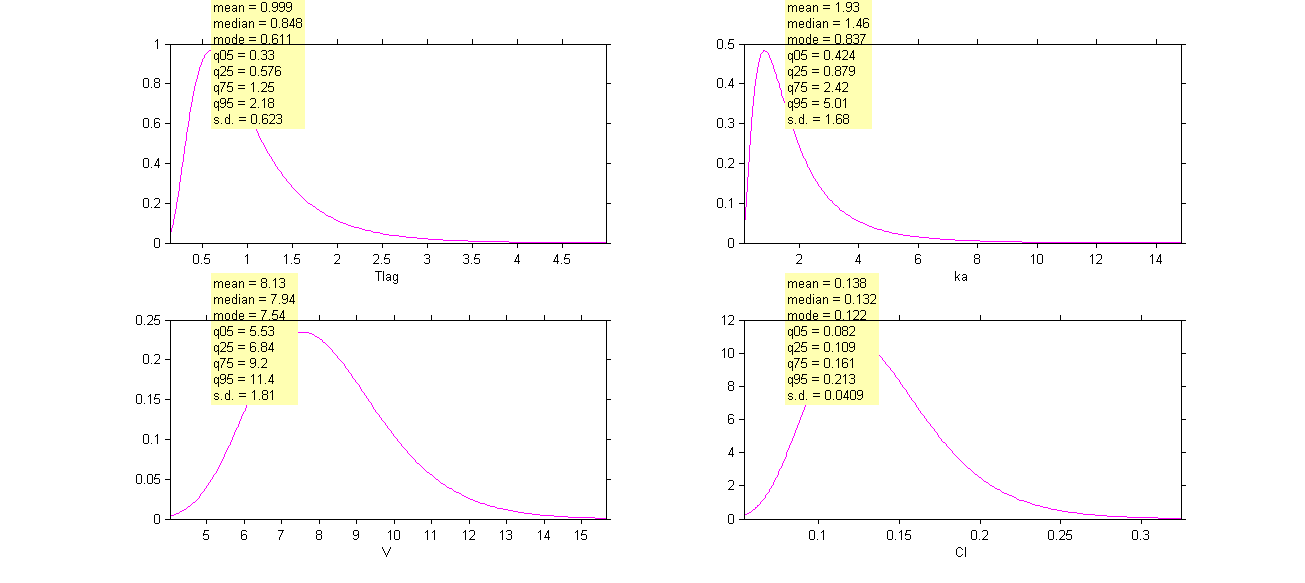
Remark 1: is not the population mean of the distribution of
, but the median of this distribution. The same property holds for the 3 other distributions which are not Gaussian.
Remark 2: here, standard deviations ,
,
and
are approximatively the coefficients of variation (CV) of Tlag, ka, V and Cl since these 4 parameters are log-normally distributed with variances < 1.
- warfarin_distribution2_project (data = ‘warfarin_data.txt’, model = ‘lib:oral1_1cpt_TlagkaVCl.txt’)
Other distributions for the PK parameters are used in this project:
- we use a uniform distribution on (0,3) for Tlag. This is equivalent to rescale a probit-normal distribution on (0,3)
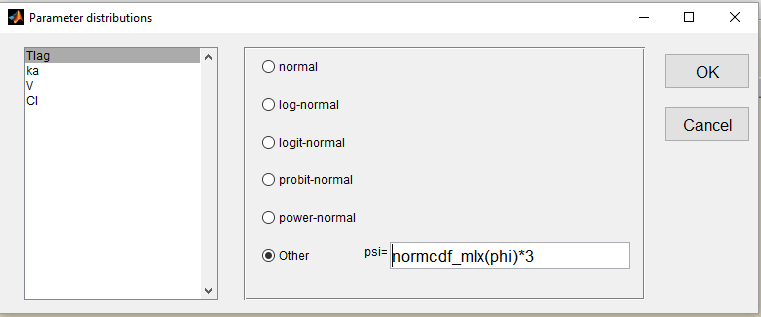
and fix the population value to 1.5 (=(0 + 3)/2) and the standard deviation
to 1:

- we use a logit-normal distribution rescaled on (0.25, 4.25) for ka:
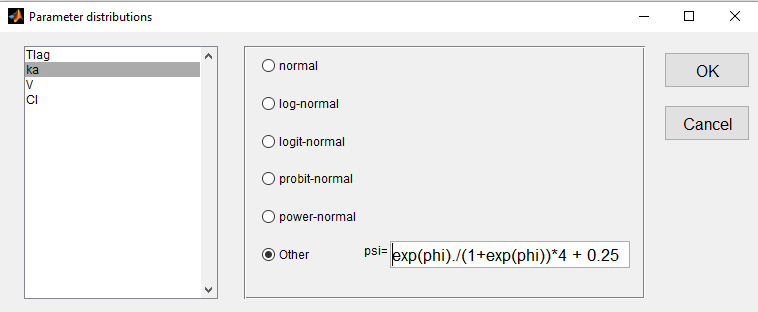
- a normal distribution for
- a log-normal distribution for
Distributions of Tlag and ka are therefore defined as Custom:
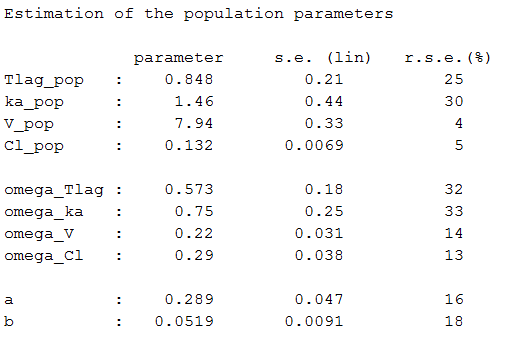
Estimated parameters are the parameters of the 4 transformed normal distributions and the parameters of the residual error model:
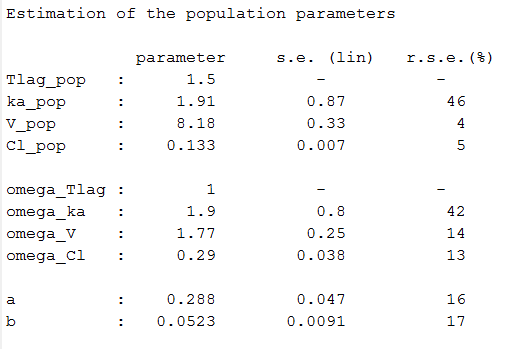
Here, and
means that
while
and
means that
The four probability distribution functions are displayed Figure Parameter distributions:
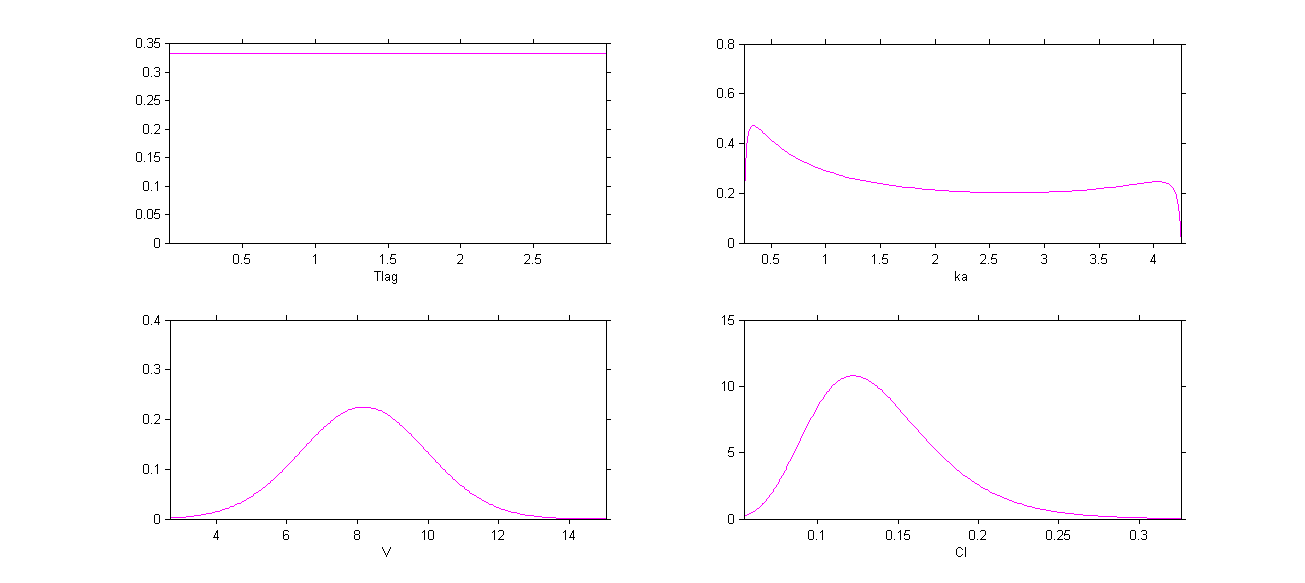
Remark 1: population values ,
,
and
are the medians of the distributions of the 4 PK parameters.
Remark 2: here, standard deviations ,
and
cannot be interpreted as coefficients of variation (CV) since Tlag, ka and V[ are not log-normally distributed.
Correlation structure of the random effects
- warfarin_distribution3_project (data = ‘warfarin_data.txt’, model = ‘lib:oral1_1cpt_TlagkaVCl.txt’)
Correlation between the random effects can be introduced in the model. A block structure is used in this project, assuming linear correlations between and
and between
and
:

Estimated population parameters now include these 2 correlations:
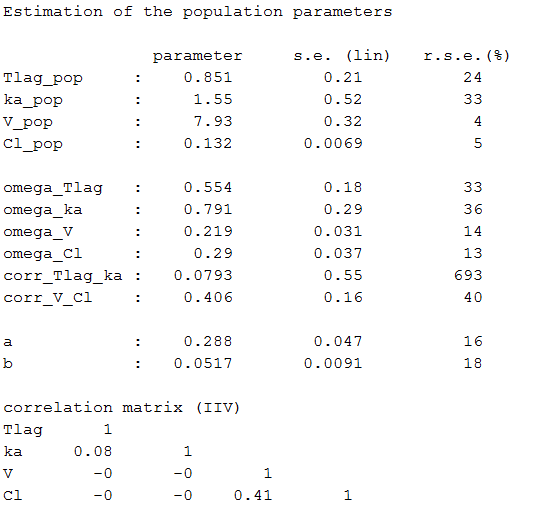
Remark: corr_Tlag_ka is not the correlation between the individual parameters and
but the (linear) correlation between the random effects
and
.
- warfarin_distribution4_project (data = ‘warfarin_data.txt’, model = ‘lib:oral1_1cpt_TlagkaVCl.txt’)
In this example, does not vary in the population, which means that
for all
, while the three other random effects are correlated:

Estimated population parameters now include the 3 correlations between ,
and
: