- The PK library
- The PD library
- Link a PK model and a PD model
- Link a PK model and a PD model with an effect compartment
- Create and use your own libraries
Objectives: learn how to use the Monolix libraries of PKPD models and create your own libraries.
Projects: warfarinPKlibrary_project, PDsim_project, warfarinPKPDlibrary_project, warfarinPKPDelibrary_project, hcv_project
The PK library
- theophylline_project (data = ‘theophylline_data.txt’ , model=’lib:oral1_1cpt_kaVCl.txt’)
A one compartment PK model for oral administration with first order absorption and linear elimination is used for fitting the theophylline data. The model oral1_1cpt_TlagkaVCl from the PK library is used:
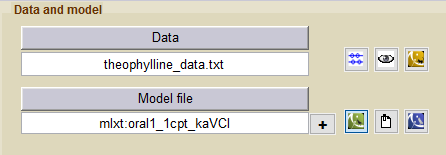
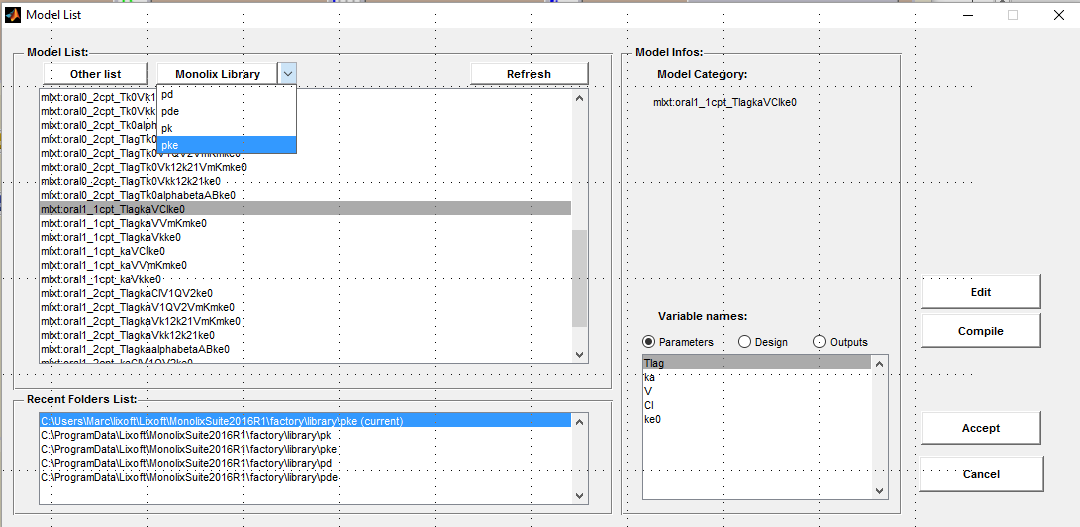
Click on the button Model file to select or modify a model. Then, select the PK library from the menu Monolix
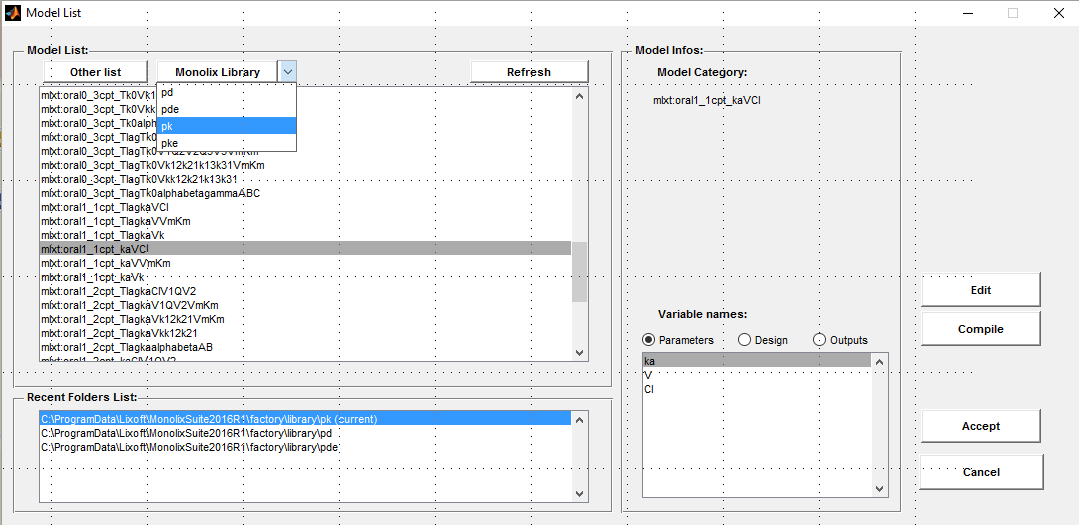
Click on the Edit button to edit the model:
[LONGITUDINAL]
input = {ka, V, Cl}
EQUATION:
Cc = pkmodel(ka, V, Cl)
OUTPUT:
output = Cc
The name of the PK model appears in the section [LONGITUDINAL] of the project file
<MODEL>
[INDIVIDUAL]
input = {ka_pop, omega_ka, V_pop, omega_V, Cl_pop, omega_Cl}
DEFINITION:
ka = {distribution=lognormal, typical=ka_pop, sd=omega_ka}
V = {distribution=lognormal, typical=V_pop, sd=omega_V}
Cl = {distribution=lognormal, typical=Cl_pop, sd=omega_Cl}
[LONGITUDINAL]
input = a
file = 'lib:oral1_1cpt_kaVCl.txt'
DEFINITION:
concentration = {distribution=normal, prediction=Cc, errorModel=constant(a)}
The PD library
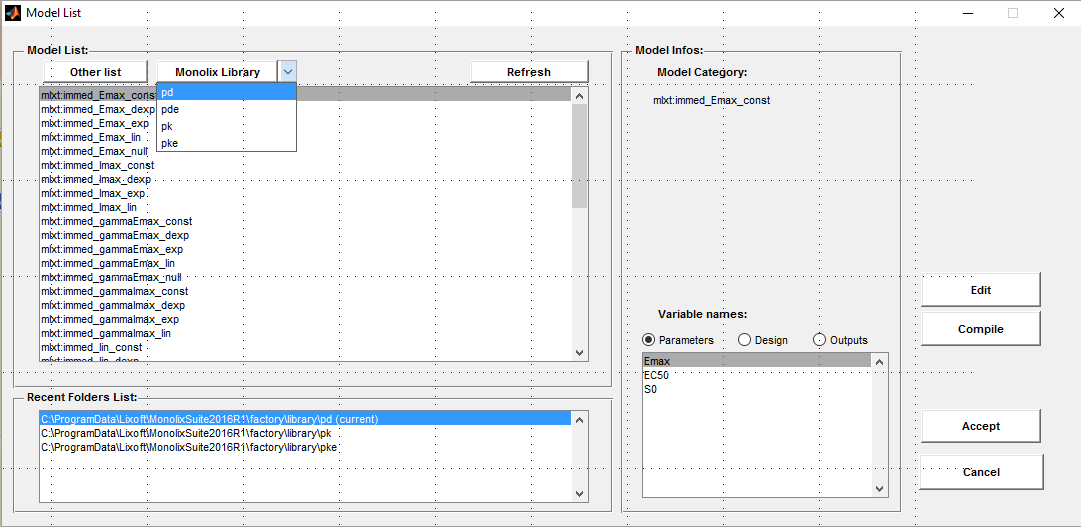
- PDsim_project (data = ‘PDsim_data.txt’ , model=’lib:immed_Emax_const.txt’)
An Emax PD model with constant baseline is used for this project.
Link a PK model and a PD model
- warfarinPKlibrary_project (data = ‘warfarin_data.txt’ , model=’lib:oral1_1cpt_TlagkaVCl.txt’)
We first fit the PK data of the warfarin data using a model from the PK library
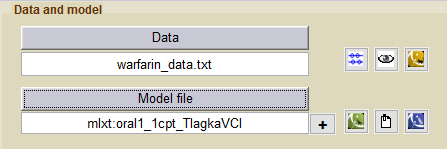
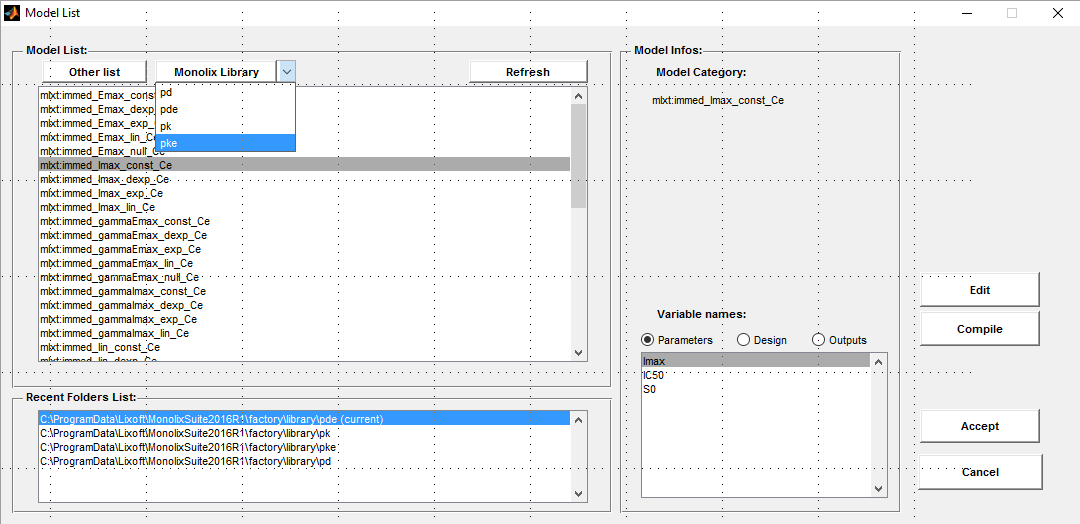
- warfarinPKPDlibrary_project (data = ‘warfarin_data.txt’ , model= {‘lib:oral1_1cpt_TlagkaVCl.txt’, ‘lib:immed_Imax_const.txt’})
To fit simultaneously the PK and the PD warfarin data, we need to link the PK model with a PD model. Click on the +
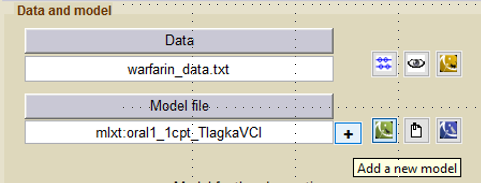
and select a model from the PD library. We will use here the immediate response model immed_Imax_const from the Monolix PD library
[LONGITUDINAL]
input = {Cc, Imax, IC50, S0}
Cc = {use=regressor}
EQUATION:
E = S0*(1 - Imax*Cc/(Cc+IC50))
OUTPUT:
output = E
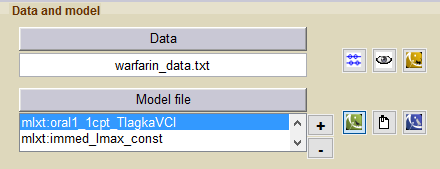
Two models then appear in the list of model files:
Link a PK model and a PD model with an effect compartment
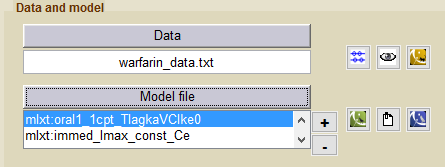
- warfarinPKPDelibrary_project (data = ‘warfarin_data.txt’ , model= {‘lib:oral1_1cpt_TlagkaVClke0.txt’, ‘lib:immed_Imax_const_Ce.txt’})
Inserting an effect compartment is made possible by using the PKe library
and the PDe library
Then, these 2 models are linked for fitting simultaneously the PK and the PD warfarin data.
Create and use your own libraries
- 8.case_studies/hcv_project (data = ‘hcv_data.txt’ , model=’VKlibrary/hcvNeumann98_model.txt’)
You can use the mlxEditor, or any other text editor, for creating new models and new libraries of models. A basic library which includes several viral kinetics model is available in 8.case_studies/model. Click on Other list for selecting this library or any other existing library of models.