Version 2016R1
This documentation is for Monolix Suite 2016R1.
©Lixoft
Monolix
Monolix (MOdèles NOn LInéaires à effets miXtes) is a platform of reference for model based drug development. It combines the most advanced algorithms with unique ease of use. Pharmacometricians of preclinical and clinical groups can rely on Monolix for population analysis and to model PK/PD and other complex biochemical and physiological processes. Monolix is an easy, fast and powerful tool for parameter estimation in non-linear mixed effect models, model diagnosis and assessment, and advanced graphical representation. Monolix is the result of a ten years research program in statistics and modeling, led by Inria (Institut National de la Recherche en Informatique et Automatique) on non-linear mixed effect models for advanced population analysis, PK/PD, pre-clinical and clinical trial modeling & simulation.
Objectives
The objectives of Monolix are to perform:
- Parameter estimation for nonlinear mixed effects models
- computing the maximum likelihood estimator of the population parameters, without any approximation of the model (linearization, quadrature approximation, …), using the Stochastic Approximation Expectation Maximization (SAEM) algorithm,
- computing standard errors for the maximum likelihood estimator
- computing the conditional modes, the conditional means and the conditional standard deviations of the individual parameters, using the Hastings-Metropolis algorithm
- Model selection and diagnosis
- comparing several models using some information criteria (AIC, BIC)
- testing hypotheses using the Likelihood Ratio Test
- testing parameters using the Wald Test
- Easy description of pharmacometric models (PK, PK-PD, discrete data) with the Mlxtran language
- Goodness of fit plots
An interface for ease of use
Monolix can be used either via a graphical interface or command lines for powerful scripting. This means less programming and more focus on exploring models and pharmacology to deliver in time. The interface can be seen here
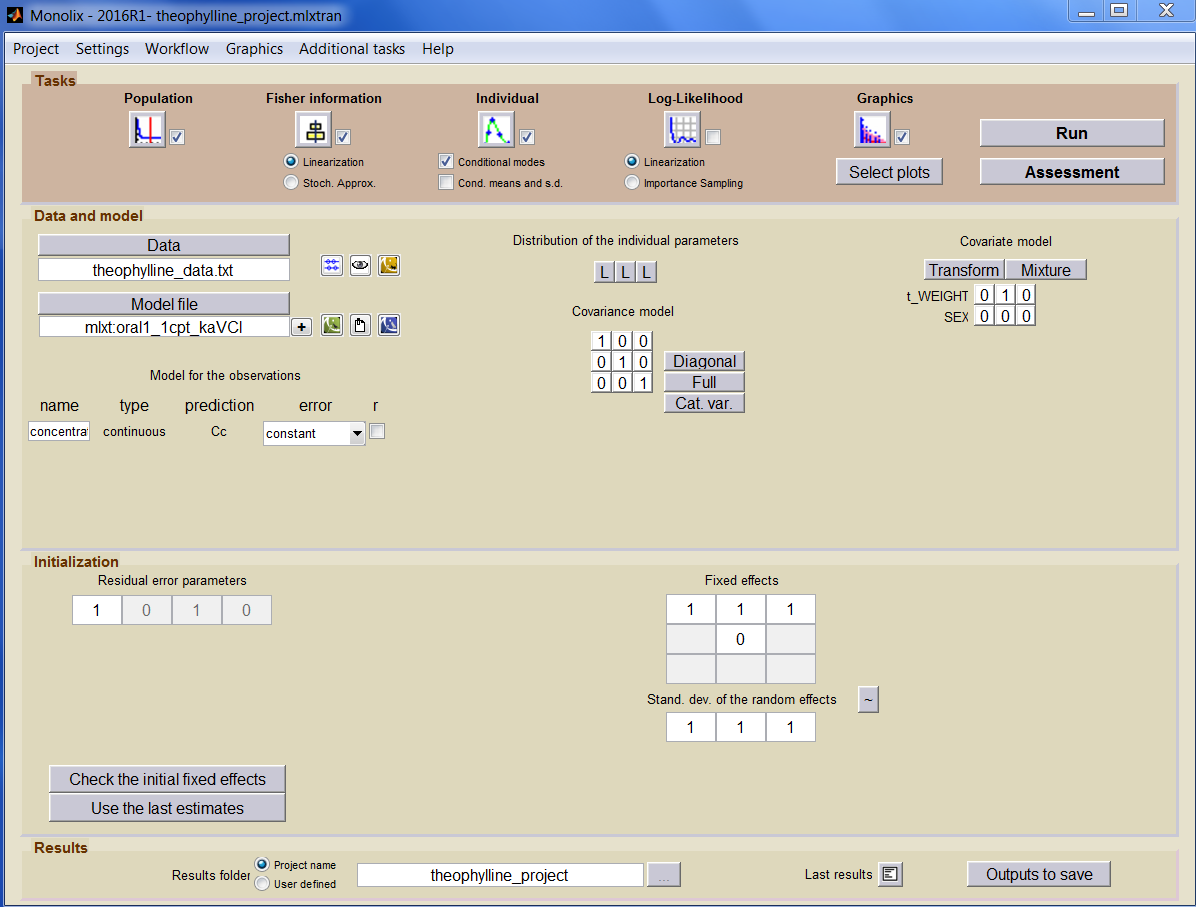
One can see several blocks in the interface
- Tasks
- Data and model
- Initialization
- Results
Each of this block has a specific section on this website. An advanced description of available graphics is also provided.