- Ordinary differential equations based model
- Don’t forget the initial conditions!
- Delayed differential equations based model
- Case studies
Objectives: learn how to implement a model with ordinary differential equations (ODE) and delayed differential equations (DDE).
Projects: tgi_project, seir_project, seir_noDelay_project
Ordinary differential equations based model
- tgi_project (data = tgi_data.txt , model = tgi_model.txt)
We consider here the tumor growth inhibition (TGI) model proposed by Ribba *et al.* (Ribba, B., Kaloshi, G., Peyre, M., Ricard, D., Calvez, V., Tod, M., . & Ducray, F., *A tumor growth inhibition model for low-grade glioma treated with chemotherapy or radiotherapy*. Clinical Cancer Research, 18(18), 5071-5080, 2012.). This model is defined by a set of ordinary differential equations
where is the total tumor size. This set of ODEs is valid for
, while
for .This model (derivatives and initial conditions) can easily be implemented with Mlxtran:
[LONGITUDINAL]
input = {K, KDE, KPQ, KQPP, LAMBDAP, GAMMA, DELTAQP, PT0, Q0}
PK:
depot(target=C)
EQUATION:
t0=0
PT_0=PT0
Q_0 = Q0
PSTAR = PT+Q+QP
ddt_C = -KDE*C
ddt_PT = LAMBDAP*PT*(1-PSTAR/K) + KQPP*QP - KPQ*PT - GAMMA*KDE*PT*C
ddt_Q = KPQ*PT - GAMMA*KDE*Q*C
ddt_QP = GAMMA*KDE*Q*C - KQPP*QP - DELTAQP*QP
Remark: t0, PT_0 and Q_0 are reserved keywords that define the initial conditions.
Then, the graphic of individual fits clearly shows that the tumor size is constant until and starts changing according to the model at
.
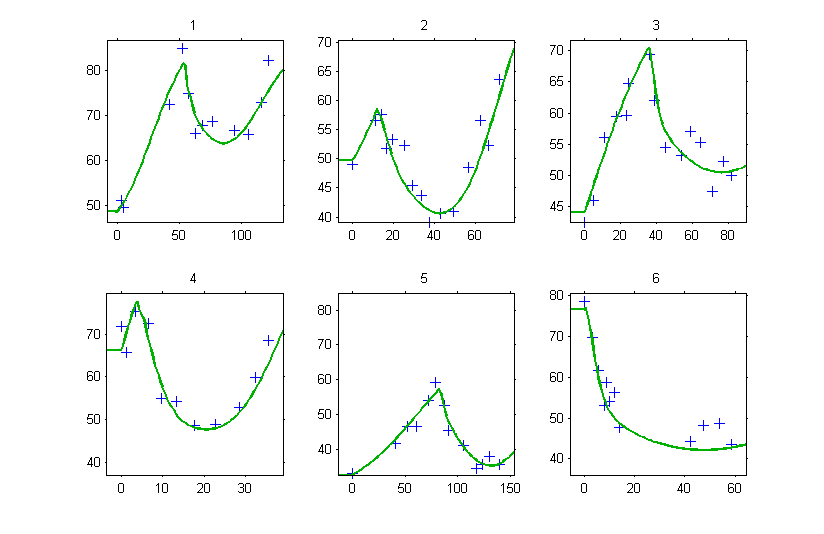
Don’t forget the initial conditions!
- tgiNoT0_project (data = tgi_data.txt , model = tgi1NoT0_model.txt)
The initial time is not specified in this example
[LONGITUDINAL]
input = {K, KDE, KPQ, KQPP, LAMBDAP, GAMMA, DELTAQP, PT0, Q0}
PK:
depot(target=C)
EQUATION:
PT_0=PT0
Q_0 = Q0
PSTAR = PT+Q+QP
ddt_C = -KDE*C
ddt_PT = LAMBDAP*PT*(1-PSTAR/K) + KQPP*QP - KPQ*PT - GAMMA*KDE*PT*C
ddt_Q = KPQ*PT - GAMMA*KDE*Q*C
ddt_QP = GAMMA*KDE*Q*C - KQPP*QP - DELTAQP*QP
Since is missing, Monolix uses the first time value encountered for each individual. If, for instance, the tumor size is computed for
for the individual fits, then
will be used for defining the initial conditions, which introduces a shift in the plot:
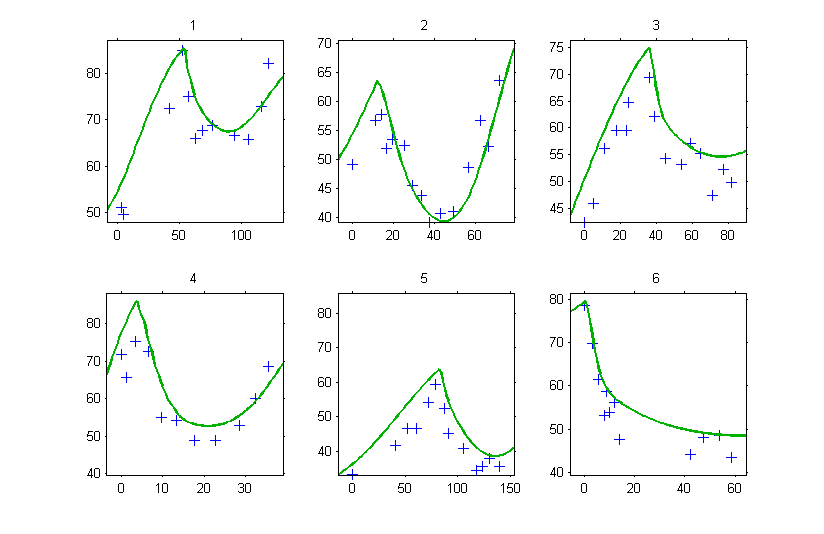
Conclusion: don’t forget to specify properly the initial conditions of a system of ODEs!
Delayed differential equations based model
A system of delay differential equations (DDEs) can be implemented in a block EQUATION of the section [LONGITUDINAL] of a script Mlxtran. Mlxtran provides the command delay(x,T) where x is a one-dimensional component and T is the explicit delay. Therefore, DDEs with a nonconstant past of the form
can be solved.
- seir_project (data = seir_data.txt , model = seir_model.txt)
The model is a system of 4 DDEs:
[LONGITUDINAL]
input = {A, lambda, gamma, epsilon, d, tau, omega}
EQUATION:
t0 = 0
S_0 = 15
E_0 = 0
I_0 = 2
R_0 = 3
N = S + E + I + R
ddt_S = A - d*S - lambda*S*I/N + gamma*delay(I,tau)*exp(-d*tau)
ddt_E = lambda*S*I/N - d*E - lambda*delay(S,omega)*delay(I,omega)*exp(-d*omega)/(delay(I,omega) + delay(S,omega) + delay(E,omega) + delay(R,omega))
ddt_I = -(gamma+epsilon+d)*I + lambda*delay(S,omega)*delay(I,omega)*exp(-d*omega)/(delay(I,omega) + delay(S,omega) + delay(E,omega) + delay(R,omega))
ddt_R = gamma*I - d*R - gamma*delay(I,tau)*exp(-d*tau)
Introducing these delays allows to obtain nice fits for the 4 outcomes, including :
- seir_noDelay_project (data = seir_data.txt , model = seir_model.txt)
On the other hand, if ODEs are used, i.e. without any delay,

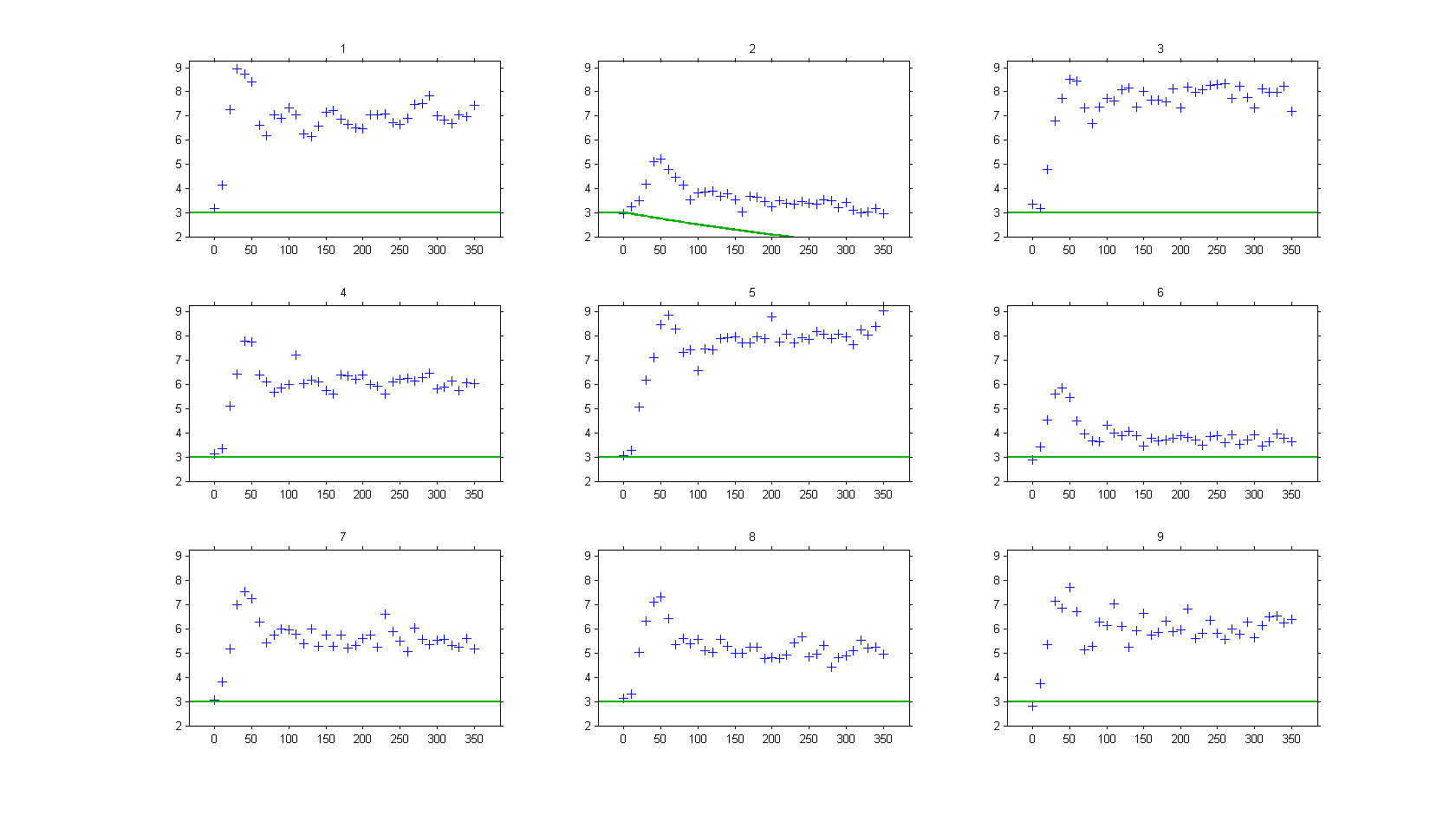
then, the model is not able anymore to fit correctly the observed data, such as :
Case studies
- 8.case_studies/arthritis_project